Comparing DNA Metabarcoding with traditional Quadrat-Based Botanical Surveys

Dr. Hannah Vallin
14 April 2025
Dr. Hannah Vallin, a postdoctoral research assistant at the Institute of Biological, Environmental & Rural Sciences (IBERS), has published her first principle-author paper in the journal Ecology and Evolution.
The study highlights the potential of DNA metabarcoding as a scalable alternative to traditional botanical surveys for assessing plant species composition in grassland ecosystems.
DNA metabarcoding is a technique used to identify multiple species within an environmental sample by analysing their DNA. It combines:
- DNA barcoding- which identifies species using short, unique genetic markers
with
- High-throughput sequencing- which allows for the simultaneous identification of many species from a single sample.
It has several potential benefits to traditional survey methods.
- It offers the capability to process multiple sequences rapidly and cost-effectively in parallel, often leading to higher taxonomic resolution
- It provides the ability to identify species throughout the growing season
- It reduces the time spent in the field
The study, titled "Comparative Analysis of Pasture Composition: DNA Metabarcoding Versus Quadrat-Based Botanical Surveys in Experimental Grassland Plots", explores the effectiveness of DNA metabarcoding compared to quadrat-based surveys under various defoliation regimes.
Traditional botanical surveys, often time-consuming and reliant on taxonomic expertise, identified 16 taxa in the experimental grassland plots. In contrast, DNA metabarcoding detected 25 taxa. Despite detecting more taxa, the study noted some discrepancies in identification, with sequence data resolving some taxa only to the genus level.
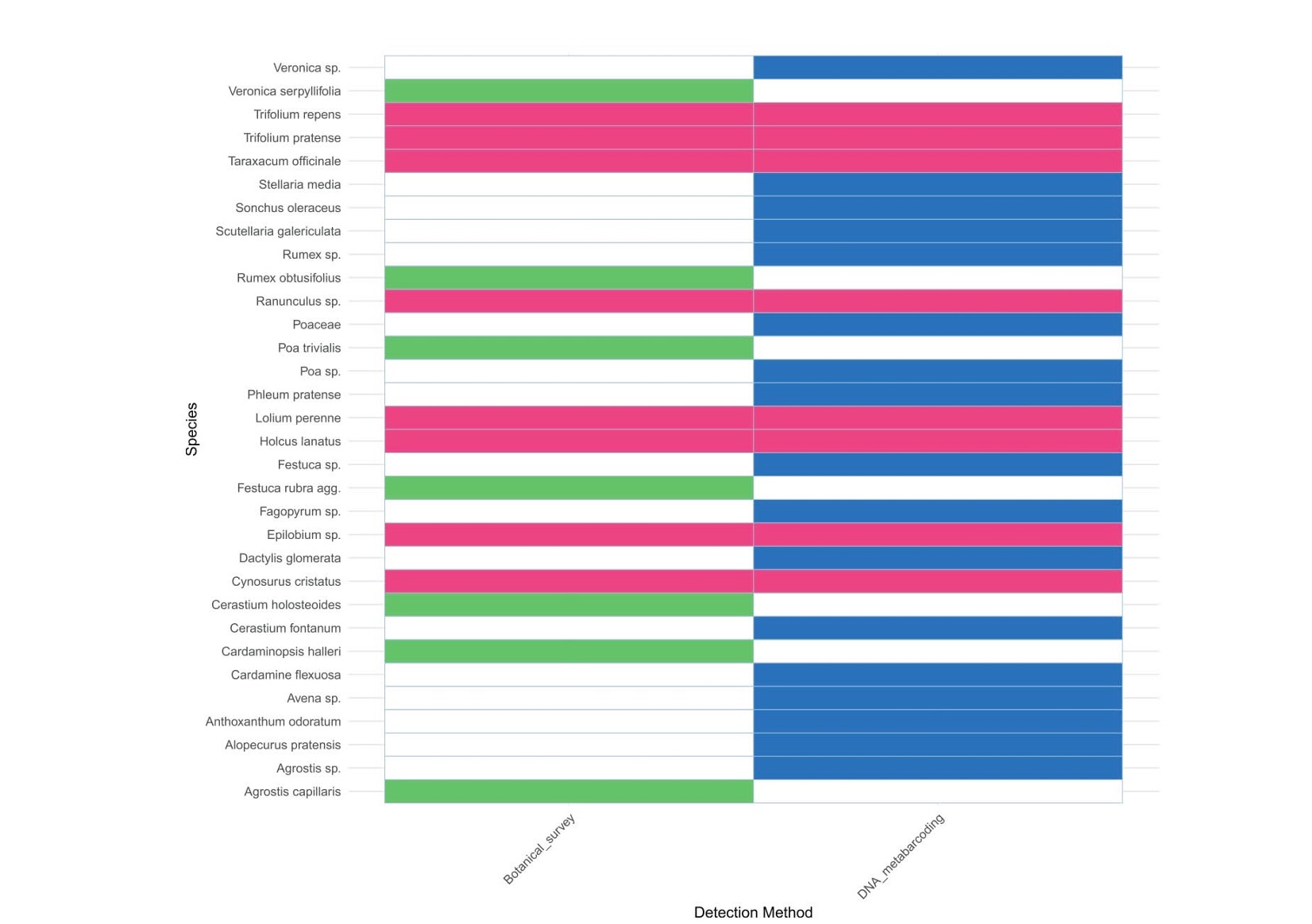
Heat table to show taxa identified in metabarcoding results (blue), botanical survey results (green) and taxa identified in both data-sets (pink).
Both methods provided comparable results, revealing statistically significant differences in species composition between treatments, with higher diversity observed in cut versus grazed plots. However, the semi-quantitative nature of DNA metabarcoding limits its capacity to accurately reflect species abundance, posing challenges for ecological interpretations where precise quantification is required.
Despite these limitations, DNA metabarcoding offers a broader view of biodiversity and can complement traditional methods, providing new opportunities for efficient biodiversity monitoring. The findings support the integration of DNA metabarcoding into biodiversity assessments, particularly when used alongside traditional techniques. Further refinement of bioinformatics tools and reference databases will enhance their accuracy and reliability, enabling more effective monitoring of grassland biodiversity and sustainable management practices.
This study underscores the value of DNA metabarcoding as a tool for understanding plant community responses to management interventions, paving the way for more efficient and comprehensive biodiversity monitoring in grassland ecosystems. Read the full paper here: https://onlinelibrary.wiley.com/doi/epdf/10.1002/ece3.71195