Proof of Principle Experiments: A Rediscovery Task
A "Rediscovery Task" was used to determine the performance of the Robot Scientist. This involved rediscovering the functions of ORFs from the well studied Aromatic Amino Acid Biosynthesis (AAA) pathway of S. cerevisiae, shown in figure 1. Of the 16 ORFs involved in the AAA pathway, only 8 were found to be auxotrophs, these were YBR166C, YDR007W, YDR035W, YDR354W, YER090W, YGL026C, YKL211C and YNL316C. Of the 22 metabolites involved in the pathway, only 9 were available for experimentation, these are shown in table 1 along with their relative costs.
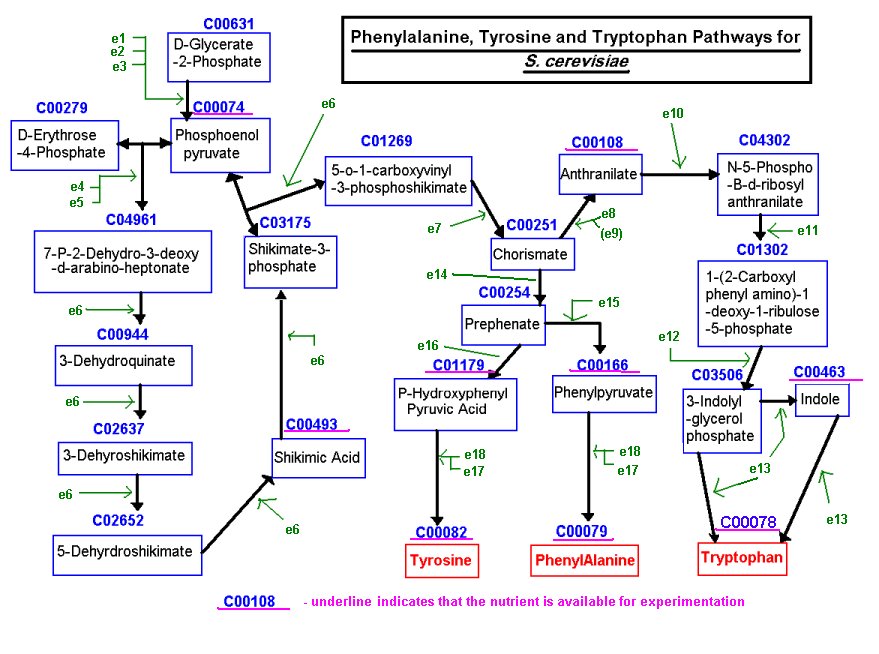
Figure 1: The Aromatic Amino Acid Biosynthesis pathway of S. cerevisiae
| Metabolite | Name | Relative Cost (Log cost) |
|---|---|---|
| C00108 | Anthranilate | 10 (1.0) |
| C00166 | Phenylpyruvate | 30 (1.48) |
| C00078 | L-Tryptophan | 53 (1.72) |
| C00079 | L-Phenylalanine | 53 (1.72) |
| C00082 | L-Tyrosine | 53 (1.72) |
| C00463 | Indole | 190 (2.28) |
| C01179 | P-Hydroxyphenyl Pyruvic Acid | 195 (2.29) |
| C00493 | Shikimic Acid | 633 (2.80) |
| C00074 | Phosphoenol Pyruvate | 9375 (3.98) |
Table 1: Relative Costs of Metabolites available for the Rediscovery Task
Knockout mutants corresponding to the 8 auxotrophic ORFs were created and made available to the robot, as were the 9 metabolites, with a concentration of 0.2 mg/l. Considering multiples of the 9 metabolites, a total of 2^9 i.e. 512 experiments are possible for each of the 8 ORFs, giving a total experiment space of 4096. However the study was limited to single and pairs of metabolites i.e. 45 combinations per ORF, or 360 possible experiments in total. The capacity of the robot was limited to considering 1 experiment for 4 ORFs per 48 hour period (c 6hrs preparation by the robot and 24+ hrs growth time for the knockouts. Exhaustive experimentation would therefore require more than 180 days, given that experiments could be interleaved: one set of experiments for 4 ORFs being prepared while experiments for the other 4 ORFs are being incubated.
However, simulations of the experimentation showed that solutions for most of the knockouts could be acquired in 5 iterations (i.e 10 days), so the maximum allowable number of experiments was set at 5 (MAXI).
As well as a proof of principle study for the Robot Scientist, these experiments were also designed to evaluate the performance of the intelligent experiment selection strategy, by comparing it with choosing experiments at random, and with a nave strategy that always chooses the cheapest available experiment. Simple algorithms for these strategies are given below:
| Intelligent Experiment Selection (ase) | Random Choice | Cheapest Experiment (naive) |
|---|---|---|
Select and execute |
I=0 |
I=0 |
where H is the set of currently valid hypotheses, S is the set of hypotheses corresponding to the solution, T is the set of currently available trials, t_min is the cheapest available trial, t_minEC is the trial that minimises the expected experimental cost, t_random is a trial randomly chosen from T, I is the current iteration and MAXI is the maximum allowable number of iterations. After a particular trial is executed by any of the strategies, that trial is removed from T.
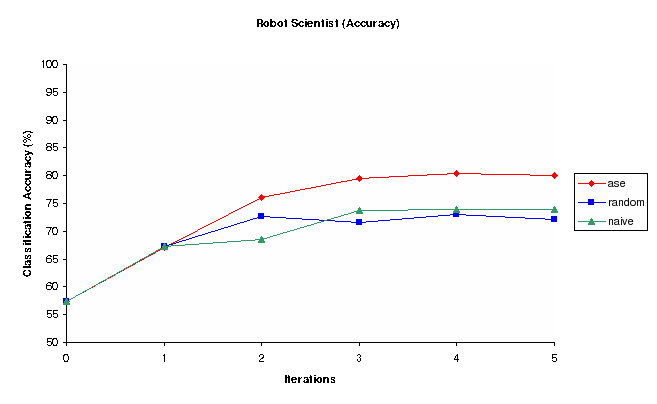
Figure 2: Classification Accuracy vs Iterations for Random, Nave and Ase Experiment selection methods (Robot)
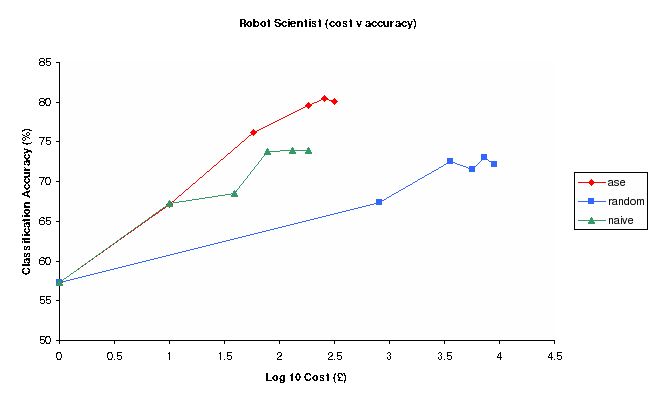
Figure 3: Classification Accuracy vs Log Experimental cost for Random, Nave and Ase Experimental selection methods (Robot)
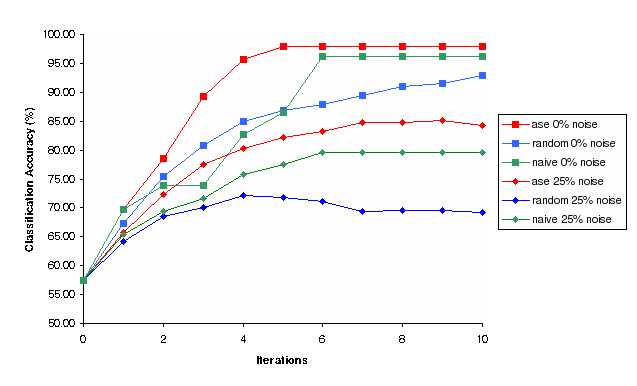
Figure 4: Classification Accuracy vs Iterations for Random, Nave and Ase experimental selection methods for 0% and 25% noise (Simulation)
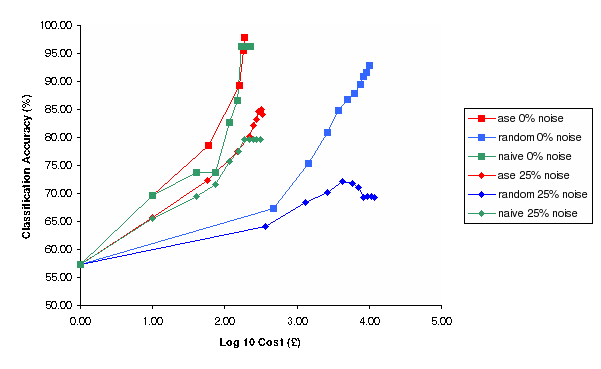
Figure 5: Classification Accuracy vs Log Cost of Experimentation for Random, Nave and Ase experimental selection methods for 0% and 25% noise (Simulation)
| Enzyme | EC Number | Enzyme Name | Yeast ORF(s) | Gene Name |
|---|---|---|---|---|
| e1 | 4.2.1.11 | phospopyruvate hydratase | YGR254W | ENO1 |
| e2 | 4.2.1.11 | phospopyruvate hydratase | YHR174W | ENO2 |
| e3 | 4.2.1.11 | phospopyruvate hydratase | YMR323W | ERR1 |
| e4 | 4.1.2.15 (now 2.5.1.54) | 3-deoxy-7-phosphoheptulonate synthase | YBR249C | ARO4 |
| e5 | 4.1.2.15 (now 2.5.1.54) | 3-deoxy-7-phosphoheptulonate synthase | YDR035W | ARO3 |
| e6 | 4.6.1.3 (now 4.2.3.4) 4.2.1.10 Unknown 1.1.1.25 2.7.1.71 2.5.1.19 |
3-dehydroquinate synthase 3-dehydroquinate dehydratase Unknown shikimate dehydrogenase shikimate kinase 3-phosphoshikimate 1-carboxyvinyltransferase |
YDR127W | ARO1 |
| e7 | 4.6.1.4 (now 4.2.3.5) | chorismate synthase | YGL148W | ARO2 |
| e8 | 4.1.3.27 | anthranilate synthase | YER090W | TRP2 |
| e9 | 4.1.3.27 | anthranilate synthase | YER090W YKL211C |
TRP2 TRP3 |
| e10 | 2.4.2.18 | anthranilate phosphoribosyltransferase | YDR354W | TRP4 |
| e11 | 5.3.1.24 | phosphoribosylanthranilate isomaerase | YDR007W | TRP1 |
| e12 | 4.1.1.48 | indole-3-glycerol-phosphate synthase | YKL211C | TRP3 |
| e13 | 4.2.1.20 | tryptophan synthase | YGL026C | TRP5 |
| e14 | 5.4.99.5 | chorismate mutase | YPR060C | ARO7 |
| e15 | 4.2.1.51 | prephenate dehydratase | YNL316C | PHA2 |
| e16 | 1.3.1.13 | prephenate dehydrogenase (NADP+) | YBR166C | TYR1 |
| e17 | 2.6.1.7 | kynurine-oxoglutarate transaminase | YGL202W | ARO8 |
| e18 | 2.6.1.7 | kynurine-oxoglutarate transaminase | YHR137W | ARO9 |
Table 2: Enzymes, ORFs and Genes that participate in the AAA pathway of S. cerevisiae
| ORF | Solution |
|---|---|
| YBR166C | e16 |
| YDR007W | e10 & e11 & e12 |
| YDR035W | e5 |
| YDR354W | e10 & e11 & e12 |
| YER090W | e8 & e9 |
| YGL026C | e10 & e11 & e12 & e13 |
| YKL211C | e9 & e10 & e11 & e12 |
| YNL316C | e15 |
Table 3: Solutions for the AAA pathway Rediscovery Task, mappings from ORF to enzyme(s). Multiple solutions indicate that the necessary metabolites for any further refutation of hypotheses were not available
